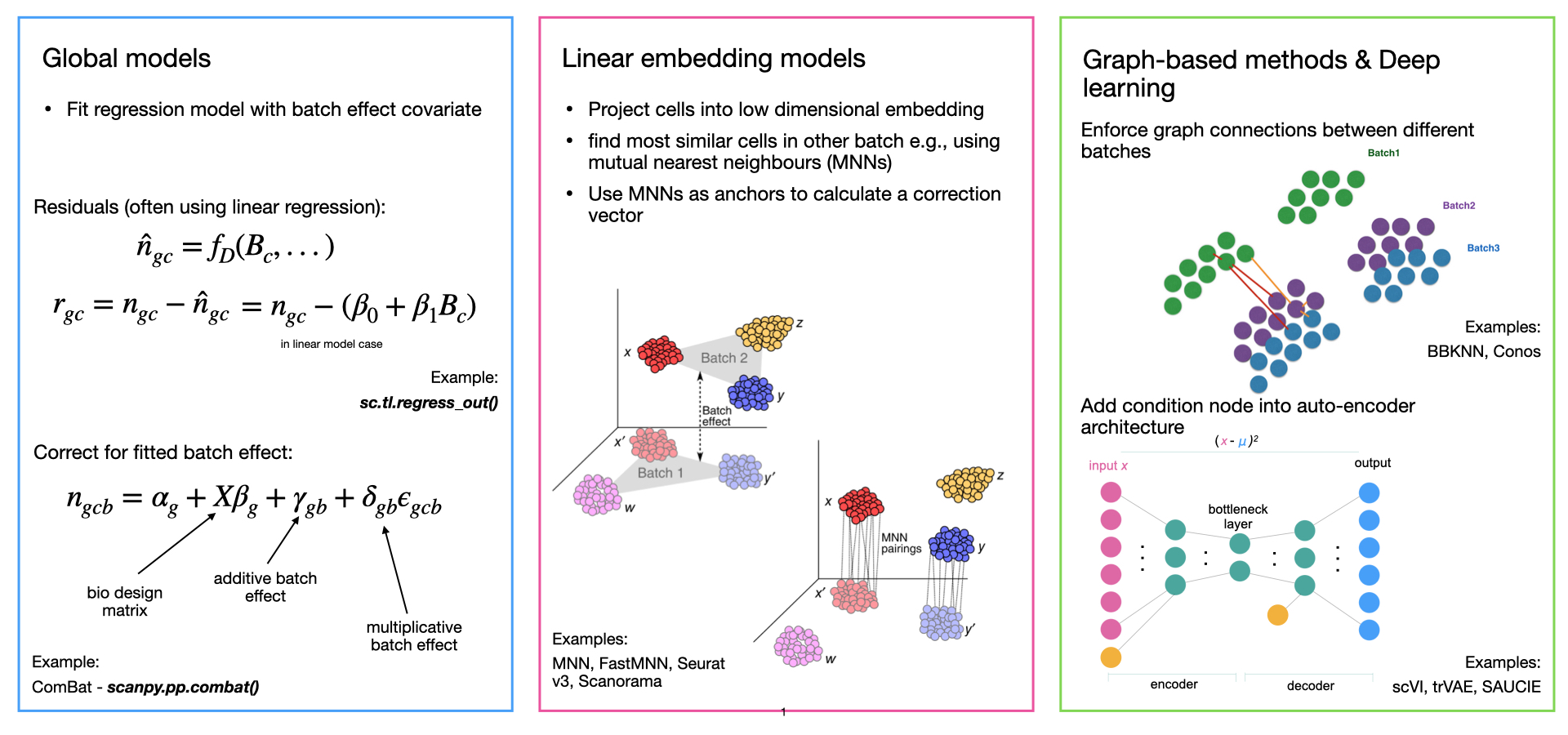
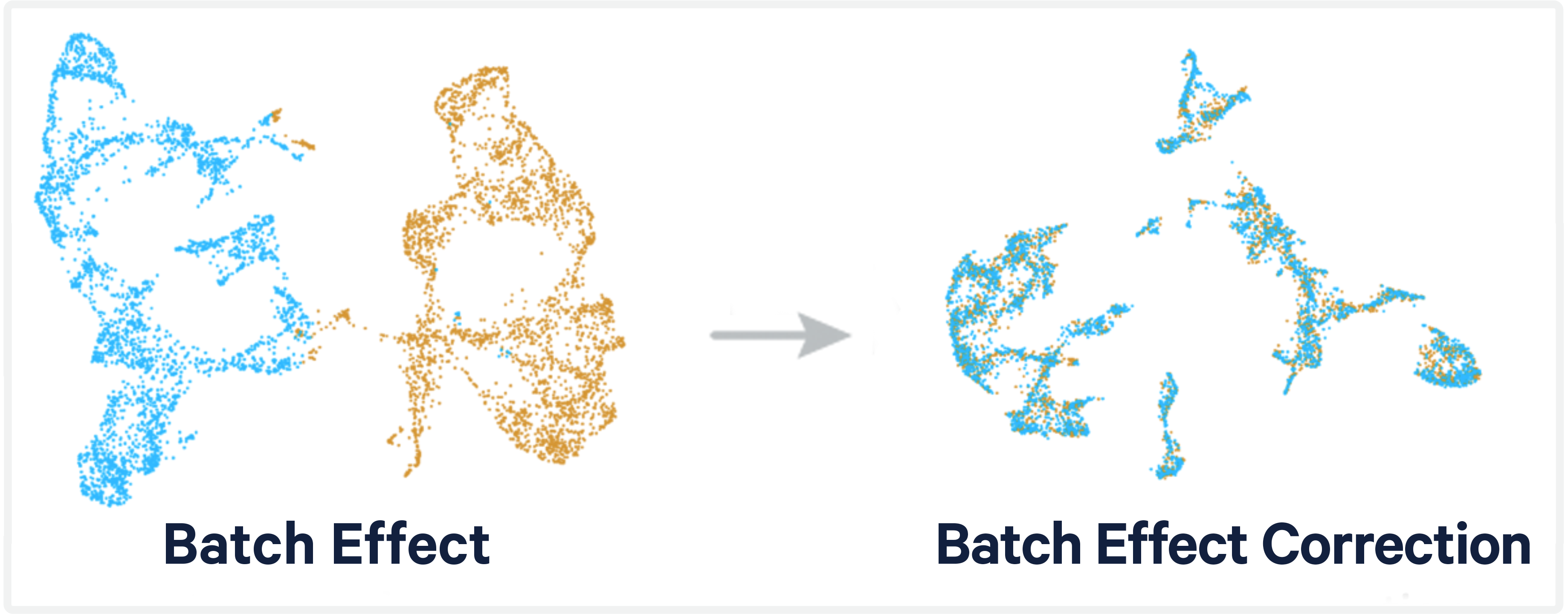
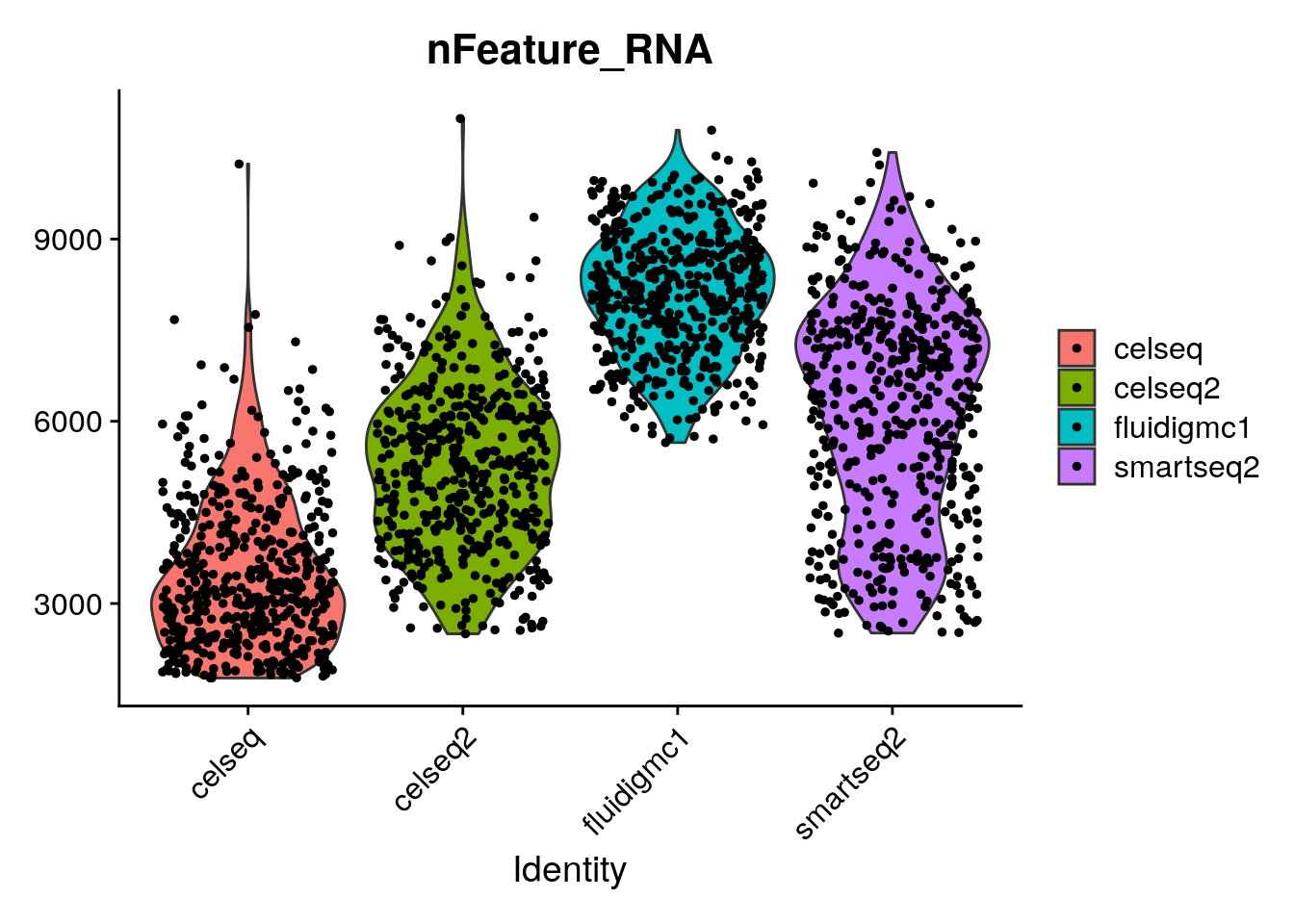
Comparison of Scanpy-based algorithms to remove the batch effect from single-cell RNA-seq data | Cell Regeneration | Full Text

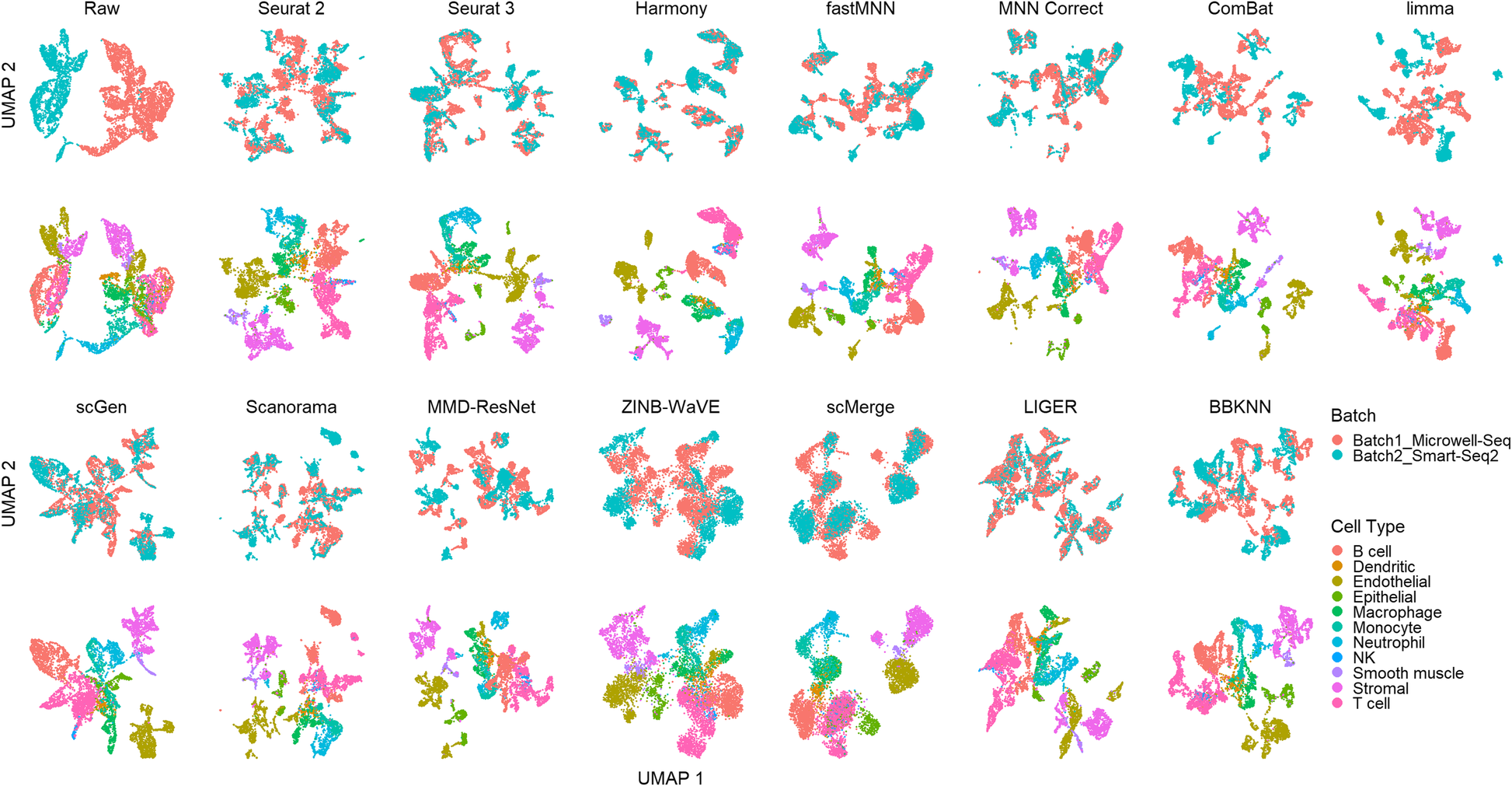
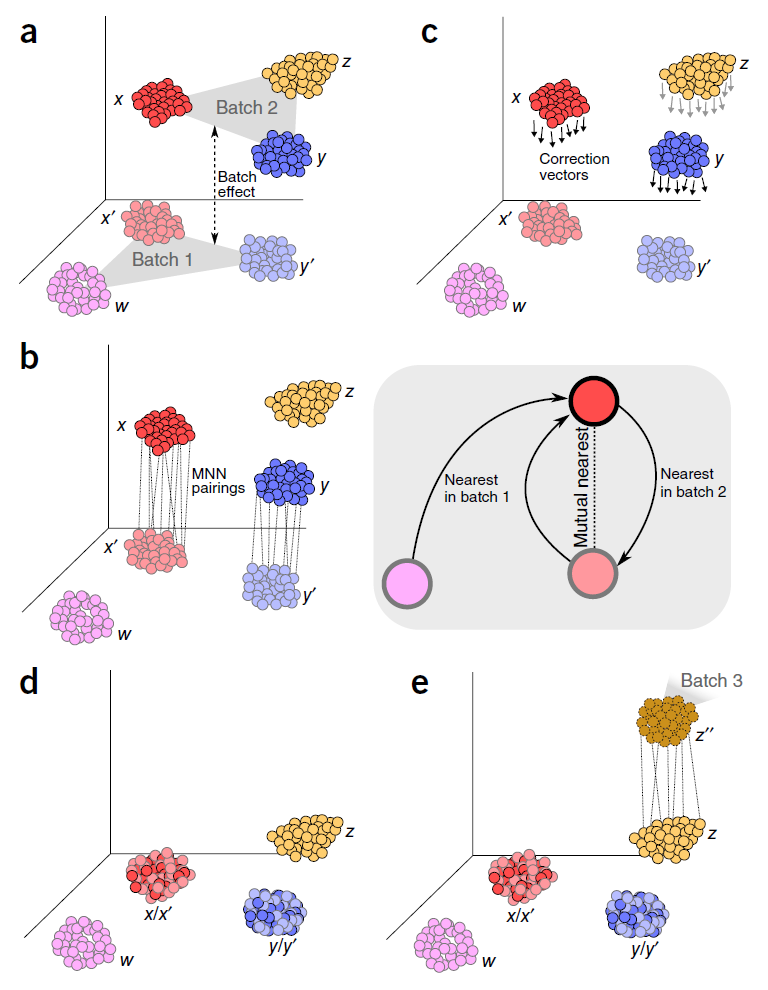
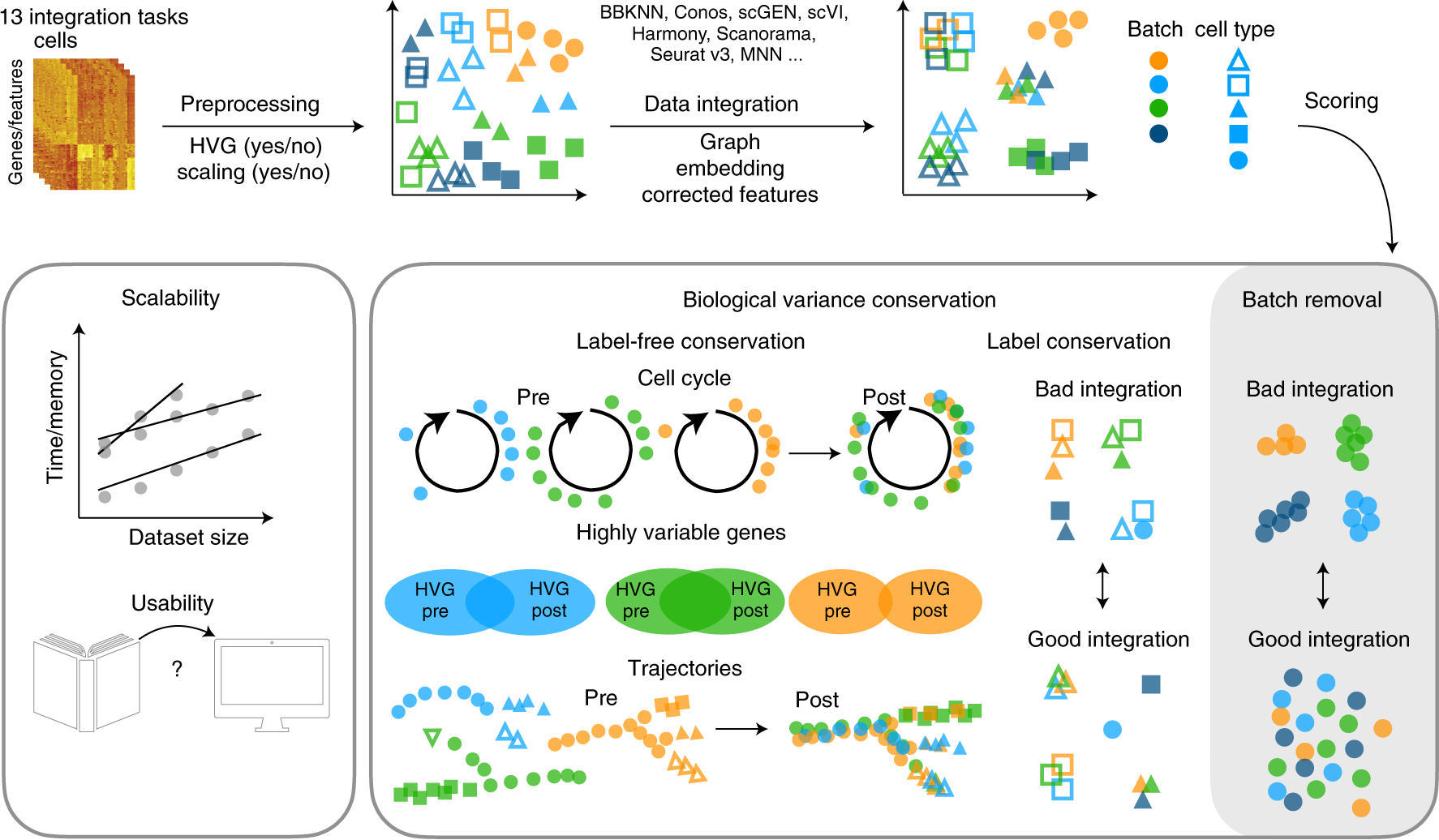
A benchmark of batch-effect correction methods for single-cell RNA sequencing data | Genome Biology | Full Text
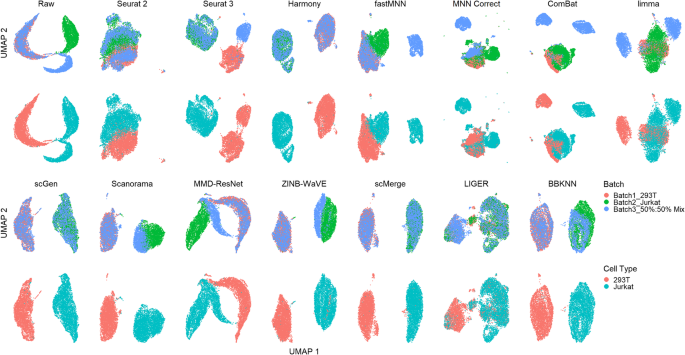
A benchmark of batch-effect correction methods for single-cell RNA sequencing data | Genome Biology | Full Text

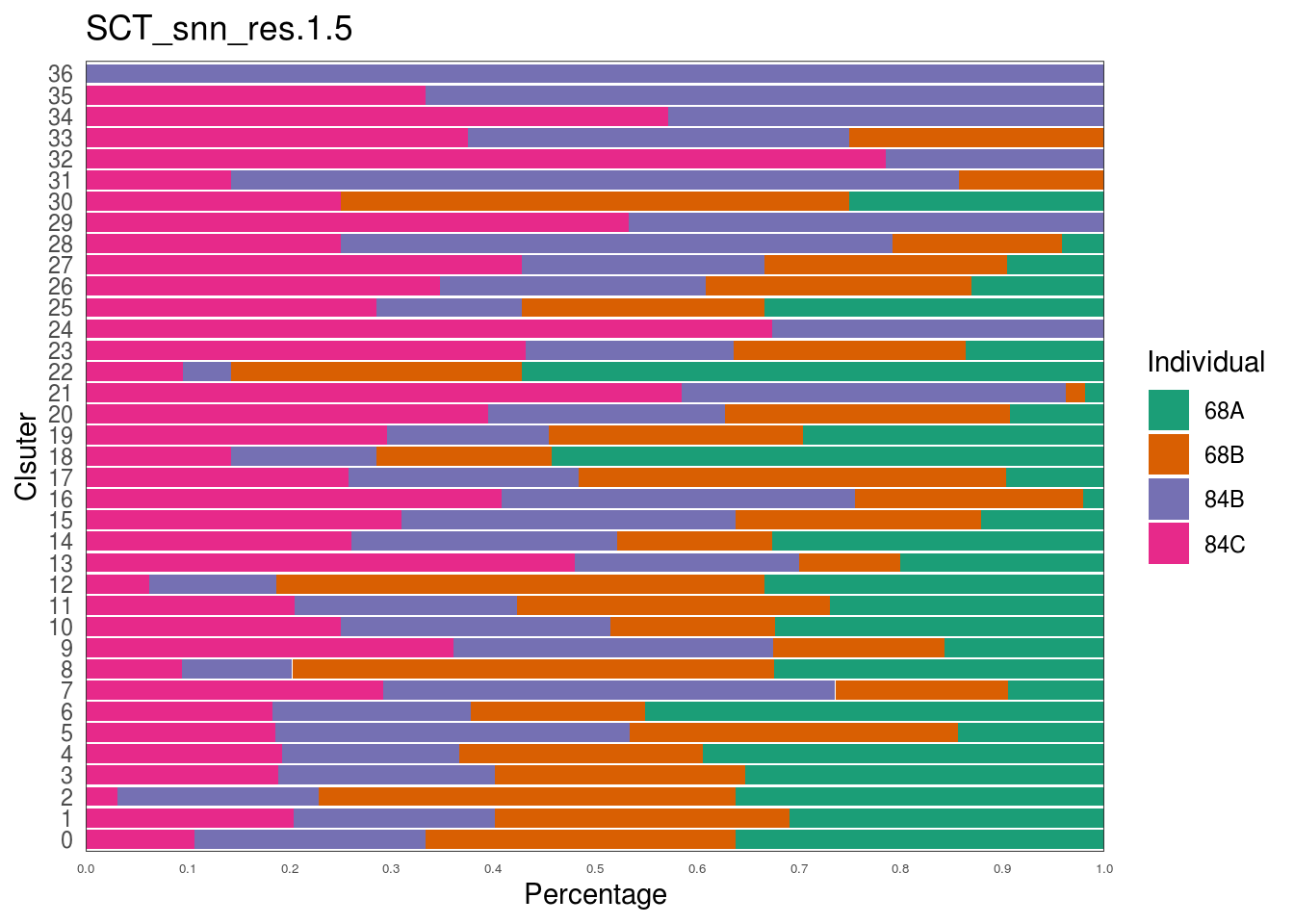
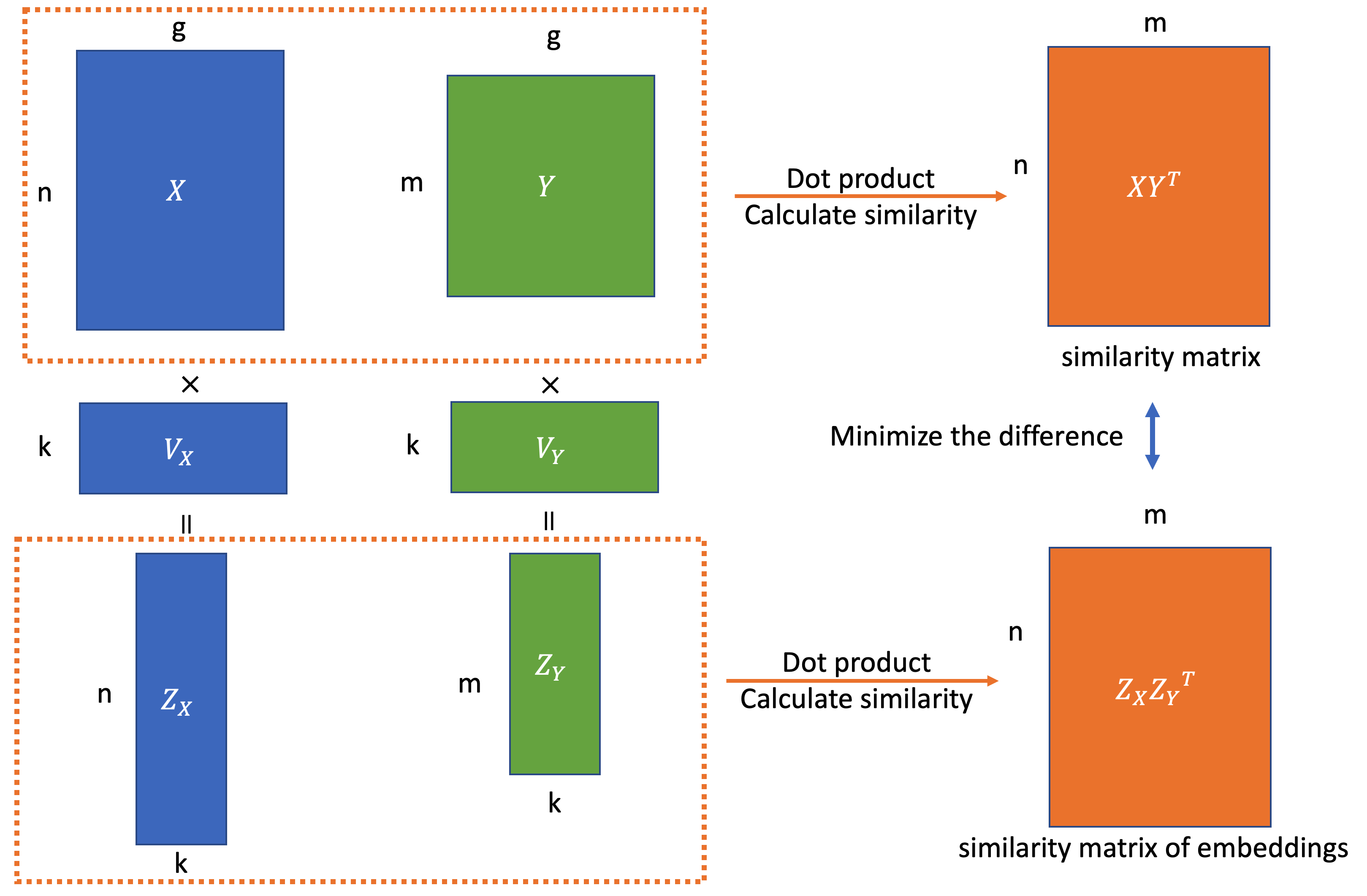
scID Uses Discriminant Analysis to Identify Transcriptionally Equivalent Cell Types across Single-Cell RNA-Seq Data with Batch Effect - ScienceDirect

Comparison of Scanpy-based algorithms to remove the batch effect from single-cell RNA-seq data | Cell Regeneration | Full Text
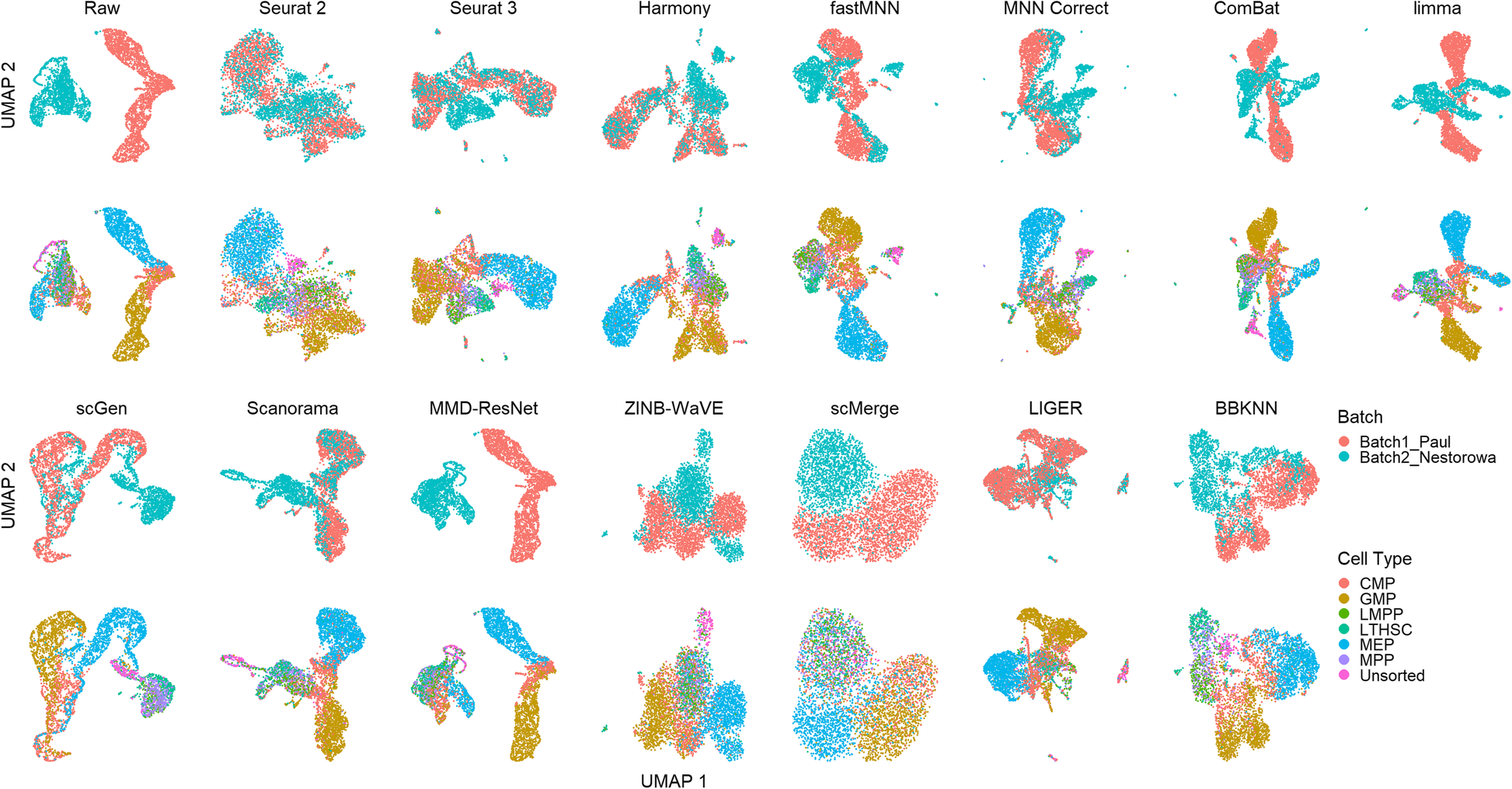
A benchmark of batch-effect correction methods for single-cell RNA sequencing data | Genome Biology | Full Text